Scientists headed by a team at the University of Tokyo have developed a new CRISPR-based gene-editing tool that they suggest could lead to better treatments for patients with genetic disorders. The tool is a version of the compact AsCas12f enzyme that incorporates mutations giving it the same effectiveness as the Cas9 enzyme commonly used for gene editing, but which is only a third of the size. The smaller size means that more of the AsCas12f enzyme can be packed into carrier viruses and delivered into cells for greater efficiency.
Through their newly reported studies, the team created a library of possible AsCas12f mutations and selected which to combine and engineer into activity-enhanced, enAsCas12f enzyme variants with 10 times more editing ability than the original unmutated enzyme. The engineered AsCas12f has been tested in mice and could potentially be used to develop more effective treatments for patients, the researchers suggested.
Study lead Osamu Nureki, PhD, at the University of Tokyo department of biological sciences, and colleagues across universities in Japan, reported on their development in Cell, in a paper titled “An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis.” In their paper, the team concluded, “Taken together, enAsCas12f variants could offer a minimal genome-editing platform for in vivo gene therapy.”
The gene-editing tool CRISPR has given researchers the ability to replace and alter segments of DNA, and scientists are applying the technology in fields as diverse as malaria prevention and crop production. In more recent years human trials have started, that aim to use CRISPR-based technology to address genetic disorders.
Two widely used genome-editing tools in human cells are SCas9 and AsCas12a, but there are limitations, the authors noted. “… their relatively large size poses a limitation for delivery by cargo-size-limited adeno-associated virus vectors.” The common way to deliver genetic material into a host cell is to use a modified virus as a carrier. Adeno-associated viruses (AAVs) are not harmful to patients, can enter many different types of cells to introduce CRISPR enzymes like Cas9, and have a lower likelihood of provoking an undesired immune response compared to some other methods. However, like any parcel delivery service, there is a size limit. Its large size means that Cas9 can lack efficiency when used for gene therapy. “Cas9 is at the very limit of this size restriction, so there has been a demand for a smaller Cas protein that can be efficiently packaged into AAV and serve as a genome-editing tool,” said Nureki.
The multi-institutional team worked to develop a smaller Cas enzyme that would prove just as active, but more efficient than Cas9. They used as their starting point AsCas12f, from the bacteria Axidibacillus sulfuroxidans. The advantage of this enzyme is that it is one of the most compact Cas enzymes found to date (422 amino acids) and less than one-third the size of Cas9. In previous tests it had shown “low but detectable” genome-editing activity in human cells, the team noted. “Therefore, AsCas12f shows great promise as a miniature genome-editing tool that can be packaged into a single AAV vector.”
Nureki and colleagues combined structural analysis and deep mutational scanning (DMS) methods to identify potential modifications to the enzyme that would improve its effectiveness in human cells. “The DMS approach combines exhaustive protein mutagenesis and functional screening with deep sequencing, enabling the assessment of the effects of thousands of mutations in a single experiment,” the authors explained. “The synergistic effects of these mutations, coupled with guide RNA [gRNA] engineering, remarkably enhanced the genome-editing efficiency of AsCas12f in human cells to levels comparable with those of both SpCas9 and engineered Cas12a effectors.”
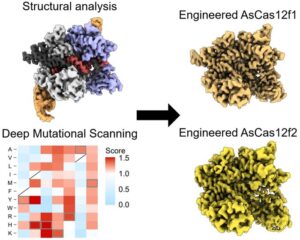
“Using a screening method called deep mutational scanning, we assembled a library of potential new candidates by substituting each amino acid residue of AsCas12f with all 20 types of amino acids on which all life is based,” Nureki continued. “From this, we identified over 200 mutations that enhanced genome-editing activity. Based on insights gained from the structural analysis of AsCas12f, we selected and combined these enhanced-activity amino acid mutations to create a modified AsCas12f. This engineered AsCas12f has more than 10 times the genome-editing activity compared to the usual AsCas12f type and is comparable to Cas9, while maintaining a much smaller size.”
The researchers commented in their paper, “The synergistic effects of these mutations, coupled with guide RNA engineering, remarkably enhanced the genome-editing efficiency of AsCas12f in human cells to levels comparable with those of both SpCas9 and engineered Cas12a effectors.”
The investigators carried out animal trials with the engineered AsCas12f system, partnering it with other genes and administering it to live mice. The encouraging results indicated that engineered AsCas12f has the potential to be used for human gene therapies, such as treating hemophilia.
The team discovered numerous potentially effective combinations for engineering an improved AsCas12f gene-editing system, and acknowledged the possibility that the selected mutations may not have been the most optimal of all the available mixes. As a next step, computational modeling or machine learning could be used to sift through the combinations and predict which might offer even better improvements.
And as the authors noted, by applying the same approach to other Cas enzymes, it may be possible to generate efficient genome-editing enzymes capable of targeting a wide range of genes. “The compact size of AsCas12f offers an attractive feature for AAV-deliverable gRNA and partner genes, such as base editors and epigenome modifiers. Therefore, our newly engineered AsCas12f systems could be a promising genome-editing platform … Moreover, with suitable adaptations to the evaluation system, this approach can be applied to enzymes beyond the scope of genome editing.”
Nureki concluded, “Elevating AsCas12f to exhibit genome-editing activity comparable to that of Cas9 is a significant achievement and serves as a substantial step in the development of new, more compact genome-editing tools. For us, the crucial aspect of gene therapy is its potential to genuinely help patients. Using the engineered AsCas12f we developed, our next challenge is to actually administer gene therapy to aid people suffering from genetic disorders.”