By Julia Milhet, Hedi Gharbi, and Venceslas Duveau, PhD
Electroencephalography (EEG) records neuronal activity inside the brain via electrodes placed on the scalp. As a real-time and dynamic method of recording ,EEG offers a direct window on neocortical electrical activity and is frequently used to monitor epileptic seizure activity and degenerative brain disorders.1
The application of machine learning has enhanced EEG utilization and the data which can be derived; as a result, its use has extended upstream from clinical applications to preclinical studies. Because brain oscillations are highly conserved properties between humans and rodents, the technology can be used to predict the efficacy of therapeutic candidates. Some of the aberrant EEG oscillations characterized in human brain disorders have also been observed in animal models.
Preclinical EEG consists of recording local field potentials in the brain of freely moving animal models; in almost any brain structure, the electrical activity of tens of thousands of neurons can be recorded, which helps clarify the underlying processes of various neurological diseases. In addition to its direct access to brain function, EEG is an objective method that does not require injection of a dye and allows for the study of dynamic processes.
Translational EEG biomarkers
Aberrant neuronal oscillatory patterns can serve as precise and nonbiased biomarkers. SynapCell has developed methods and associated validation processes to qualify aberrant EEG patterns as disease-specific biomarkers for use in compound testing. As a quantitative measure of neuronal activity, EEG offers dynamic and robust predictive results and avoids observer bias. Aberrant EEG patterns need to meet several key criteria to qualify as biomarkers:
• They need to be well documented in the clinic and found in relevant animal models (for example, hippocampal paroxysmal discharges in focal epilepsy).
• They must be modulated or reversed by reference drugs (for example, beta oscillations
in Parkinson’s disease suppressed by levodopa).
• They must be reproducible over time to confirm robustness (as determined by blinded, clinical-like latin-square designs, additional pharmacological challenges, and deeper statistical analyses).
The Cue® in vivo platform encompasses high-level modeling methods, precise recording techniques, and deep-learning based signal processing to enable quantification of brain activity in healthy and pathological conditions. The high time resolution offered by EEG recordings enables assessments of the pharmacodynamics of pharmaceutical compounds on clinically relevant models using EEG biomarkers (Figure 1).
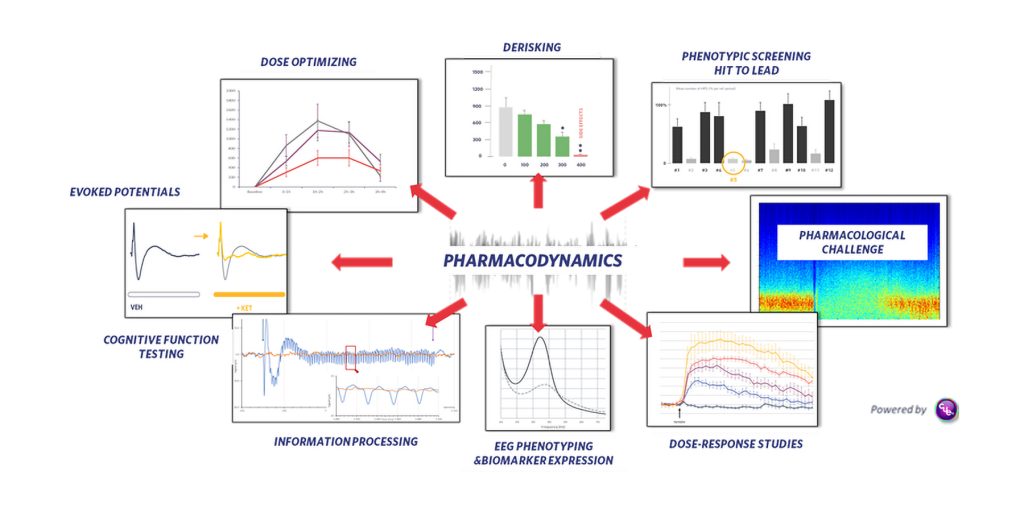
Cue accepts EEG input and supports a wide range of applications such as monitoring drug-induced oscillatory changes to determine whether a compound reaches its target and crosses the blood-brain barrier (pharmacological EEG); testing cognitive enhancers using auditory steady state response; monitoring antiepileptic properties of compounds; or evaluating antiparkinsonian as well as antidyskinetic drugs. The platform can also enable the benchmarking of drugs; the uncovering of mechanisms of action; the screening of small libraries of compounds; and the flaging of possible unwanted side effects.
The Cue platform can accommodate various dosing paradigms, including acute, subchronic, and chronic dosing paradigms, and it can be used with more complex protocols such as protein expression driven by a gene therapy. Overall, Cue provides objective decision-enabling endpoints to document the effects of test compounds on various brain diseases and can also be applied to characterize animal models (EEG phenotyping).
EEG phenotyping
The Cue platform can be used to phenotype preclinical animal models of central nervous system (CNS) disorders. Subtle oscillatory changes can be detected in a given model when compared to wild-type controls to highlight a specific EEG phenotype linked to the disease. Once highlighted, the EEG phenotype can be challenged with appropriate pharmacology to identify potential pharmacosensitivity. This process enables functional exploration using the EEG phenotype as a proxy of the disease or mechanism studied, as well as target validation and engagement with the ability to correlate a genotype (knock-in or knockout, homozygous, heterozygous) to an EEG phenotype. This feature could be used to stratify subpopulations (for example, by correlating mutations on different loci with different disease symptoms), to clarify disease mechanisms, and to support more precise and personalized treatment approaches. EEG phenotyping represents an important advance in drug discovery, as it augments the translational value of an animal model, highlighting a functional EEG biomarker of brain pathology.
An unmet need in epilepsy treatment
Although convulsive seizures experienced by epilepsy patients are well addressed by medications currently on the market, mesio-temporal lobe epilepsy (mTLE), a form of focal epilepsy, remains difficult to treat, making it a high unmet need. Most pharmacological treatments have no effect on these nonconvulsive seizures, with 30% of patients being drug refractory. A major obstacle to development of possible therapies for mTLE was the fact that none of the technologies available on the market could highlight a pathological, nonconvulsive phenotype on the mTLE mouse model.
To help find a possible treatment for mTLE, SynapCell incorporated a relevant model using the Cue platform to support the development of mTLE therapies (Figure 2). The model recapitulates a wide range of characteristics observed in human mTLE including disease semiology, histopathological alterations, EEG data, and pharmacology.2 When SynapCell’s Cue platform was used to characterize hippocampal paroxysmal discharges in this model, they were shown to be very similar to those observed in patients, as well as pharmacosensitive to the same reference drugs tested. The mTLE mouse model therefore allows prediction of drug efficacy as early as the preclinical stage.

Conclusion
The Cue platform takes advantage of the translational properties of EEG to closely replicate human disease in relevant animal models. Combined with relevant data analysis, EEG bridges the gap between preclinical research and clinical practice by accurately measuring the therapeutic potential of compounds in the brain. The platform can be applied to different therapeutical strategies, including adeno-associated virus–induced gene therapies. Benefiting from the latest technological innovations, the platform enables a more robust application of quantitative, predictive EEG measurements in CNS drug discovery.
Julia Milhet is marketing communications coordinator in medical technology, Hedi Gharbi is head of global marketing, and Venceslas Duveau, PhD, is head of sales and product development at SynapCell.
References
1. Sutter R, Kaplan PW, Schomer DL. Historical Aspects of Electroencephalography. In: Schomer DL, Lopes da Silva FH, Eds. Niedermeyer’s Electroencephalography: Basic Principles, Clinical Applications, and Related Fields. 7th ed. Oxford University Press; 2017: 1–19. DOI:10.1093/med/9780190228484.003.0001.
2. Duveau V, Roucard C. A Mesiotemporal Lobe Epilepsy Mouse Model. Neurochem. Res. 2017; 42(7):1919–1925. DOI: 10.1007/s11064-017-2239-3.
Within SynapCell, Julia Milhet is communications manager, Hedi Gharbi is head of marketing, Venceslas Duveau is head of scientific sales. Contact: [email protected]