Sponsored content brought to you by
Why Microscoop?
Spatial proteomics aims to reveal the locations of a group of proteins in cells or tissues to provide a spatial framework for the understanding of protein interactions or molecular mechanisms of biological processes. However, image-based spatial proteomics techniques used in recent advances require fluorescently labeled antibodies of proteins from prior studies, enabling mapping but limiting the capability of discovering novel protein players at a specific location of interest.
Syncell’s Microscoop™ is a game-changing platform that captures the proteome at your region of interest—no protein candidate list is needed in advance. Ultimately, the Microscoop enables hypothesis-free subcellular spatial discovery, providing users with a list of known and novel proteins.
The underlying principle behind Microscoop is its high-precision photolabeling that “microscoops” up proteins at specific subcellular regions for proteomic analysis. Another novel feature of Microscoop is its AI-assisted model, which recognizes subcellular targets more accurately than conventional computer vision based–imagevision-based image processing. The AI algorithm is applied to images of thousands of fields of view, one field of view at a time, fully automatically, to control the illumination locations for targeted photolabeling. With Microscoop, spatially specific proteins from hundreds of thousands of individual cells can be rapidly and precisely labeled and isolated to achieve the sensitivity of mass spectrometry.
Advantages of Microscoop
Microscoop, which can be applied to cell and tissue samples, captures proteins from specific subcellular structures in fixed cells and tissue biospecimens, including cryosections and formalin-fixed paraffin-embedded (FFPE) tissue specimens. Microscoop is ideal for both basic and clinical research and has immediate applications in oncology, immunotherapy, immunology, neurobiology, infectious disease, and developmental biology. Furthermore, Microscoop’s AI algorithm is ideal for the pattern recognition of a variety of selected subcellular structures, including the nucleus, immune synapses, nucleoli, stress granules, neurons, and microvilli.
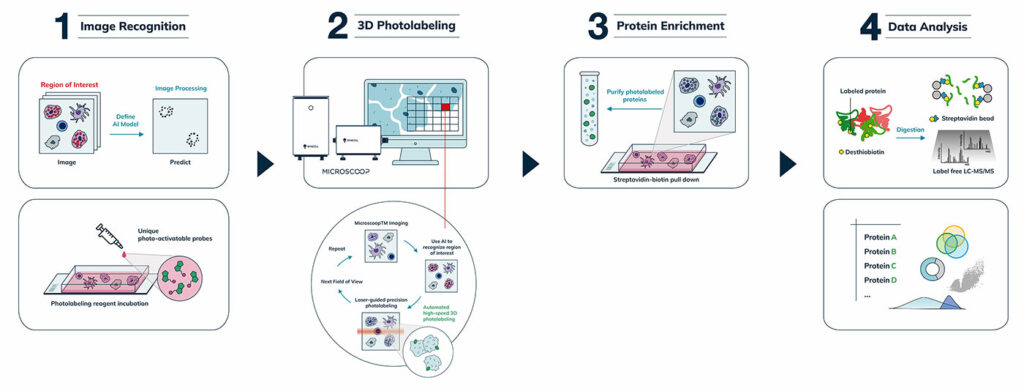
Microscoop is Transforming Science
Syncell’s Microscoop has paved the way for several pioneering studies for spatial proteomics. For example, Microscoop has been used to unveil novel protein players associated with β amyloids. The aggregation of amyloid-β peptides (Aβ) is a hallmark of Alzheimer’s disease.
Although the subcellular distribution of these peptides is well recognized under a microscope, their pathological and physiological functions remain unclear. In a recent study using Microscoop, researchers were able to biotinylate proteins in Aβ1–42 aggregates, which is a hallmark of Alzheimer’s disease, in a differentiated SH-SY5Y cell model.
Novel Protein Players
Intriguingly, the measured proteome was found to contain novel protein players that were not previously thought to be Aβ-related. The researchers further validated these newly identified proteins using antibody staining and confirmed that the proteins colocalized with Aβ1–42. Identifying these novel proteins paves the way for new diagnostic biomarkers and drug targets for Alzheimer’s disease.
Microscoop has also been used to uncover the proteome of stress granules. Stress granules—which are hard to analyze due to their multiple subcellular locations—are dynamic, non-membrane-bound assemblies of protein and RNA formed in response to cellular stress. Because these structures appear to have pathological implications in cancer and neurodegenerative disease, their composition is of great interest for therapeutic applications.
Previous proteomic studies on stress granules have been achieved through biochemical fractionation or proximity labeling before mass spectrometry (LC-MS/MS). With Microscoop, more than 10 novel stress granule proteins that have not been reported in the literature were identified and validated by immunostaining. The results also show the high specificity of the identified proteome, where 48 of the top 50 ranked proteins identified with Microscoop were known or validated as stress granule proteins. Further studies of these novel proteins can shed light on underlying mechanisms of stress granule formation and their associated functional effects.
Microscoop Is the Future
Syncell’s Microscoop enables the high specificity and high precision of proteome discovery without labor-intensive procedures. The platform’s clever way of spatial binning to accumulate enough proteins for mass spectrometry has already helped researchers identify novel proteins associated with specific subcellular compartments and structures. Identifying these novel proteins paves the way for new biomarkers or druggable targets to treat a variety of diseases, including, cancer, neurodegenerative diseases, and many others. It is expected to be broadly used for diverse biological problems with its smart scooping capability.
References
- TOTAL-SYNC ULTRA-CONTENT MICROSCOPIC OPTO-BIOTINYLATION ENABLES HIGH-SENSITIVITY HYPOTHESIS-FREE IN SITU SUBCELLULAR PROTEIN DISCOVERY. SYNCELL’S AMERICAN ASSOCIATION FOR CANCER RESEARCH (AACR) POSTER. 2023. ABSTRACT 5638. ORLANDO, FLORIDA.
- DE NOVO SPATIAL PROTEOMIC PROFILING OF IMMUNE SYNAPSES USING MACHINE LEARNING-GUIDED MICROSCOOP. SYNCELL’S AMERICAN ASSOCIATION FOR CANCER RESEARCH (AACR) POSTER. 2022. ABSTRACT 3875. NEW ORLEANS, LOUISIANA.
- MICROSCOOP, A DISCOVERY-BASED IMAGE-GUIDED PROTEOMICS TECHNOLOGY, REVEALS NOVEL FACTORS ON AMYLOID-B AGGREGATES IN DIFFERENTIATED SH-SY5Y CELLS. SYNCELL’S AMYLOID-BETA POSTER. NEUROSCIENCE 2022. SAN DIEGO, CALIFORNIA.
- NOVEL STRESS GRANULE PROTEINS IDENTIFIED BY MICROSCOOP, A MICROSCOPE-GUIDED HYPOTHESIS-FREE SUBCELLULAR PROTEIN DISCOVERY PLATFORM. SYNCELL’S STRESS GRANULES POSTER. CELL BIO 2022. WASHINGTON, DC.

Learn more syncell.com