Like Pac-Man frantically gobbling up pixelated dots, the world is furiously consuming data. Unlike Pac-Man, the world is in danger of becoming full—and soon. Data is entering our lives and driving our decisions like never before. According to Domo, a cloud software company, in every minute of 2019, Americans were using 4,416,720 gigabytes of internet data, including data for 188,000,000 emails, 4,497,420 Google searches, and even 1,400,000 Tinder swipes. Market research from We Are Social offers an international perspective: In 2019, the internet welcomed more than one million new users every day.
Whether from the internet, digital photos, or even secret military operations, data streams are flowing ever more prodigiously. Demand for data storage will soon outstrip the availability of memory-grade silicon. Additionally, current storage devices have a limited lifespan and relatively low density. However, Mother Nature has been solving this problem for about a billion years. She’s already created nature’s oldest and most reliable hard drive . . . DNA!
Because it is so dense, a single gram of DNA can store 215 petabytes (215 million gigabytes) of data. DNA is also very robust. Scientists recently obtained DNA from the tooth of a rhinoceros that lived 1.7 million years ago. They also extracted DNA from the 5000-year-old remains of Ötzi, the so-called Iceman, identifying living Austrian relatives and determining he was lactose intolerant.
But before you can store your family photos on a DNA hard drive, there are a number of challenges that must be overcome such as cost and speed of DNA synthesis. GEN spoke with several industry experts to gain insights into how this new revolution hopes to solve the global data explosion.
DNA-writing enzymes
Converting digital code to DNA code is relatively straightforward if you can navigate the world of algorithms as well as DNA synthesis and analysis. First, convert a digital file into binary and then create an algorithm using the four nucleotides of DNA to encode the information. For example, encrypt a binary digital file into a DNA data file using combinations of the binary digits 0 and 1 to represent the four bases of DNA (for example, 00 for adenine, 01 for cytosine, 10 for guanine, and 11 for thymine). After DNA synthesis and sequencing, the file is decoded. Early successes include storing a cat’s photograph, pop videos, and even Wikipedia.
“Four years ago, we had not considered DNA data storage in our five-year company plan,” muses Michael J. Kamdar, president and CEO, Molecular Assemblies. “However, the government has been a big driver to challenge the industry to move forward in solving this issue.”
Employing DNA to pack and store data will require some major advances since the current method of synthesizing DNA is expensive, labor intensive, and produces hazardous waste. “The traditional phosphoramidite chemical method of synthesis appeared in 1981 and has spawned an entire industry,” notes J. William “Bill” Efcavitch, PhD, Molecular Assemblies’ CSO. “While the costs are slowly decreasing, this method is restricted and can only produce a strand of about 100 nucleotides.”
Storing megadata will require revolutionizing DNA methods. To address this challenge, Molecular Assemblies developed a new method of economically synthesizing de novo DNA using a template-free polymerase (terminal deoxynucleotidyl transferase, TdT). The company can produce a strand that is many thousands of nucleotides long with a minimum of mechanical steps using aqueous reagents. “However, it’s the design of the nucleotide monomers that provides the ‘secret sauce,’” quips Efcavitch.
The company provided a proof-of-principle demonstration of its methodology by successfully encoding a short text message into DNA, subsequently producing it and translating it via next-generation sequencing. According to Efcavitch, “We now want to scale up the process. For example, if we need to code terabytes of information, numerous DNA strands (for example, 1012) must be simultaneously synthesized in a quick and cost-efficient manner.”
Kamdar believes that we are on the cusp of this data revolution: “While progress is certainly being made, it will take a lot more capital to accomplish these lofty goals. We are still in the early days of DNA data storage and need bigger data-oriented companies such as Amazon, Google, and Microsoft to provide the capital necessary to move this forward.”
Perfectly imperfect
Henry Lee, PhD, co-founder and CEO of Kern Systems, believes perfection is highly overrated, at least for the field of DNA-based storage. He summarizes Kern’s novel approach as follows. “DNA-based storage does not require writing DNA with single-base precision or accuracy, so we can use rapid enzymatic synthesis that generates nonperfect sequences. We developed a kinetically controlled method that uses TdT enzyme to catalyze synthesis of customized DNA strands with short homopolymeric extensions. To limit the number of bases added at each step, we use apyrase, which also competes with the polymerase for nucleotides. We then apply error-correction software to ensure full data recovery from redundantly synthesized DNA strands.”
The company also does something that breaks with tradition. Instead of utilizing binary code (that is, bits), it uses a ternary system (that is, trits) to represent information. Lee explains, “We decided to store information in transitions between nonidentical bases to reduce the need for perfect synthesis. Because there are three possible transitions between nonidentical nucleotides (for example, A to C, G, or T), the use of trits maximizes information capacity. During sequence analysis, we compact the sequence by treating all repeated nucleotides as a single occurrence and only read the transition between nucleotides for information storage. For example, the sequence ‘AAAATTTGGGGCC’ is read as ‘ATGC’ and contains three transitions: A to T, T to G, and G to C. Unique transitions are used to represent information.”
One challenge Kern faced was how to store perfect data while relying on nonperfect DNA synthesis. To do this, the company devised a codec to efficiently retrieve information from a population of imperfectly synthesized DNA strands. The codec contains error-correction schemes that are widely employed not only for genomics (that is, DNA sequencing) but also for digital communications such as cell phones and the internet.
Kern will next tackle scale-up toward megabytes and beyond. According to Lee, this will require novel hardware engineering for parallelization and automation as well as further optimization of biochemistries. “For data storage, we must improve our ability to synthesize diverse DNA sequences by orders of magnitude, compared to existing processes,” he declares. “Our ultimate goal is to synthesize at incredible scale with tunable accuracy. We are now building the foundation for large-scale enzymatic synthesis to support the demand for data storage as well as for rewriting entire genomes.”
Scale up, price down
Making long-term DNA data storage accessible and commercially viable will require not only modernizing the DNA synthesis process but also concomitantly reducing costs. Twist Bioscience recently received a large portion of a $25 million project awarded by the Intelligence Advanced Research Projects Activity (IARPA) through its Molecular Information Storage (MIST) program. The company will focus on ramping up DNA synthesis strategies as a subcontractor to the Georgia Tech Research Institute, which is the primary awardee.

Emily M. Leproust, PhD, co-founder and CEO, reflected, “We currently utilize silicon chip technology that can synthesize 1 million different DNA sequences that are printed for ~ $1000 per megabyte. Our goal is to drive down cost to $100–$1000 per gigabyte, a 1000-fold decrease.”
To accomplish this task, the company’s army of engineers will be enlisted. Leproust points out, “We combine the talents of hardware engineers, mechanical engineers, chemical engineers, software engineers, algorithm engineers, and others to build the machines and instrumentation of the future. We want to shrink the typical silicon chip of 50 microns down to 1 micron and even submicrons while at the same time packing in even higher amounts of information.”
Leproust compared the process to the evolution of transistors that, over time, squeezed more and more processes and information into transistor chip technology. But the process doesn’t happen overnight. She reports, “For an integrated system using silicon chips and semiconductor design, testing follows an 18-month cycle. We are in the early stages at the moment.”
Rise of molecular electronics
Roswell Biotechnologies is also a member of the IARPA’s MIST program led by Georgia Tech Research Institute. “Our task is on the retrieval or readout of digital data that has been stored in DNA,” comments Roswell CSO Barry Merriman, PhD. “In biomedical sequencing, we have achieved the $1000 genome, but this program wants the $1 genome. The new IARPA program is a four-year program to demonstrate the feasibility of terabyte-scale DNA storage. It is also by far the largest and most focused effort for this to date. Essentially, we are transitioning from science fiction to investment.”
The overarching mission at Roswell is to develop the first-ever molecular electronics sensor chip platform. Merriman summarizes, “Our ‘killer app’ is an electronic sensor chip that reads DNA. We are the only company pursuing a molecular electronics solution for reading DNA, that is, a single-molecule, all-electronic DNA reading sensor, deployed onto a scalable complementary metal oxide semiconductor (CMOS) chip device.”
The company is also looking to the far future for its technologies. Merriman predicts, “Our technology was specifically architected to have a very long roadmap of improved scaling (and related cost reductions) by fully leveraging the scaling available in both current and future CMOS chip foundries. As a result, we envision a not-too-distant future in which a whole-genome sequencing test in medicine will cost much less than $100, and exabyte-scale data will be stored in DNA instead of magnetic tape.”
Push-button/walk-away DNA data storage
Moving DNA-based storage to the next level will require bridging the digital and biological worlds. That is, software and hardware engineering must be combined with biology in a fully integrated and automated system. Such a system is being developed by Codex (formerly SGI-DNA). In fact, the company asserts that it has built the world’s first fully automated DNA data storage system.
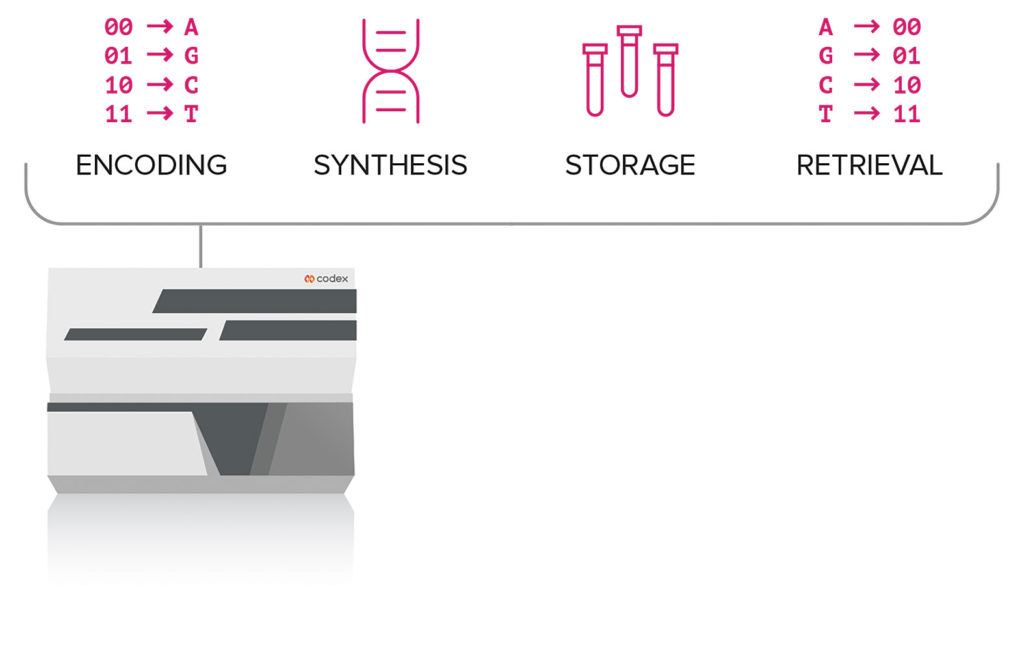
“We overcame two main challenges in developing the system,” states Codex CEO Todd R. Nelson, PhD. “The first was to integrate and automate data storage. The second was to ramp up writing of data into DNA from kilobytes per day to megabytes per day. We continue to work on a prototype using novel microfluidics engineering solutions that will enable an entire server farm of information to be stored in a single tube of DNA.”
The company’s current digital DNA storage system is an automated, miniaturized technology with a workflow that can be completed in eight hours featuring a capability of writing nearly one million base pairs of DNA per day per system. Initially, digital files are sent through the company’s portal and encoded to DNA files using proprietary algorithms. A button is pushed selecting the desired process (writing, storing, and/or retrieving files), and then the DNA data files are generated on the system.
 Codex indicates that it is the early phases of its new technology and that it has big plans for the future. Nelson discloses, “We want to create near-infinite capacity, massively parallel running systems to increase encoding, writing, and retrieval speeds. We’re working toward dramatically accelerating writing speeds on a single instrument by more than one million-fold to 10 megabytes per second or greater.”
Codex indicates that it is the early phases of its new technology and that it has big plans for the future. Nelson discloses, “We want to create near-infinite capacity, massively parallel running systems to increase encoding, writing, and retrieval speeds. We’re working toward dramatically accelerating writing speeds on a single instrument by more than one million-fold to 10 megabytes per second or greater.”
Although the field of DNA-based storage is rapidly evolving, it is likely still several years away from prime time. Nelson predicts, “I expect to see some practical applications within about three to five years. There is a huge long-term market potential in DNA storage technologies, and we will continue to invest and invent to make that future a reality.”



